Overview: What is TRIC-seq?
TRIC-seq (Total RNA Interactome Capture) is an in situ, genetics‑free proximity‑ligation method that maps native RNA–RNA contacts across bacterial transcriptomes. It preserves cellular context and resolves both intramolecular (structure) and intermolecular (regulatory/proximity) contacts at high resolution and specificity.
Each chimeric read corresponds to one ligation event between two RNA fragments that were nearby in the cell. Aggregating millions of such events resolves:
- RNA structures (Hi‑C–like self‑contact maps, identifying functional domains, insulators, and structural hot-spots).
- Regulatory networks (identifying highly connected sRNA–mRNA pairs, regulatory sponges, and trans-acting hubs).
- System‑level patterns (uncovering modular interactomes, extensive mRNA-mRNA contacts, and condensate‑like organization, driven largely by non-ribosomal transcripts).
Biological Insights
By circumventing biases associated with probe-based or UV-crosslinking strategies, TRIC-seq provides an unbiased look into the spatial orchestration of the transcriptome. Key findings unlocked by TRIC-seq include:
- Pervasive mRNA-mRNA architecture: Transcripts don't act alone; they frequently overlap geographically, participating in interconnected communities and operon-spanning regulatory cross-talk.
- Target discovery: Novel trans-acting RNA-RNA interactions can be discovered systematically by mapping robustly enriched intermolecular loops (significant Odd Ratios).
- Tertiary folds in vivo: Long-range intramolecular contacts pinpoint 5′UTR/3′UTR interactions that gate functional expression.
Using this dataset
The explorer provides pre‑loaded TRIC‑seq datasets for multiple bacteria (e.g., E. coli, Staphylococcus aureus, Stutzerimonas stutzeri, Myxococcus xanthus). Choose a preset in each tool to load data instantly.
Key fields
- Interaction count (io): unique chimeric reads supporting a pair.
- Odds ratio (Of): enrichment relative to a null; higher means more specific pairing.
- Adjusted P/FDR: multiple‑testing–corrected significance (especially for long‑range contacts).
- Feature types: 5′UTR, CDS, 3′UTR, sRNA, tRNA, housekeeping RNA (hkRNA), sponge.
- TRIC‑seq simultaneously captures intramolecular structure signals and intermolecular regulatory contacts; interpret diagonal‑proximal self‑contacts as structure and distal cross‑locus contacts as pairing/regulatory proximity.
- Contact specificity is better reflected by Of (and FDR) than by counts alone—use both when ranking candidates.
- Use long‑range (kb‑scale) enrichment to flag distal regulation, then validate in pairMAP.
GSE305265 when using this explorer.Tool guide: globalMAP
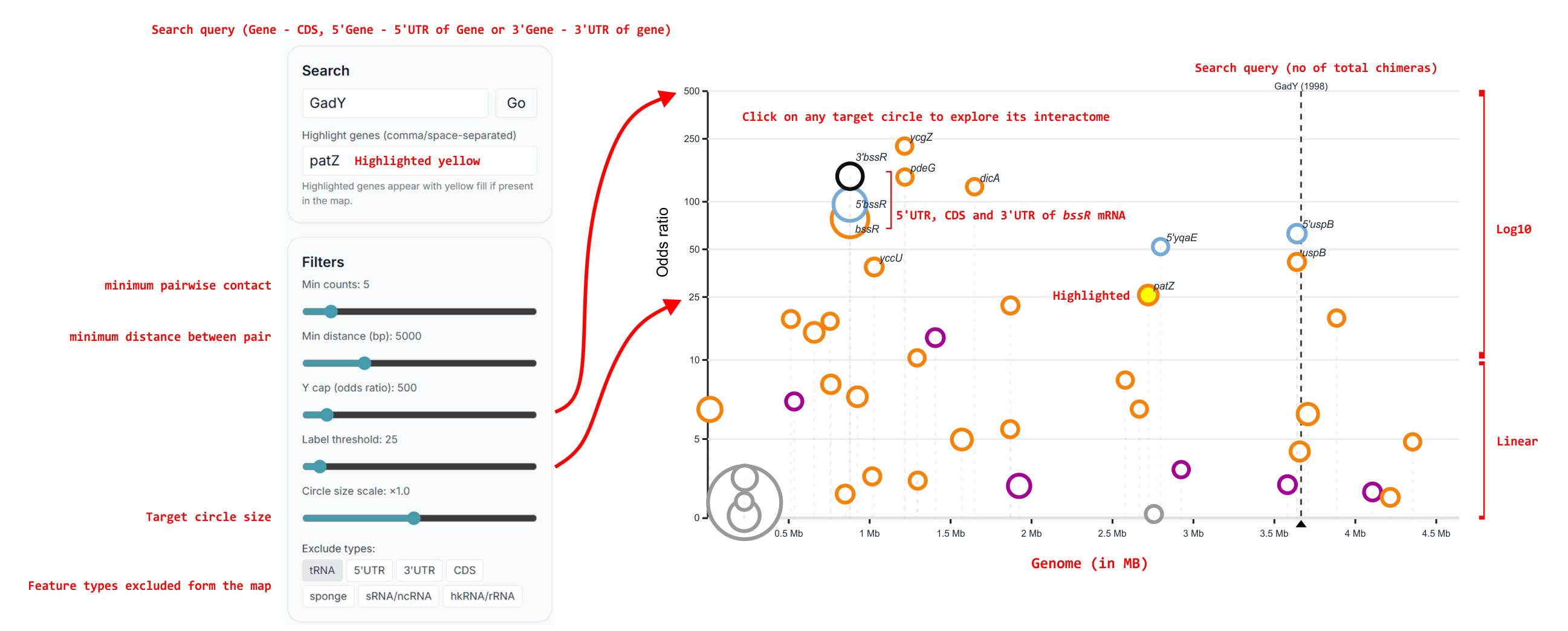
What it shows: a genome‑aware, RNA‑centric map of all partners for a selected RNA. Points encode partner position, Of, io, and feature type. The table includes quick links (↗) to external gene databases based on your selected annotation preset.
How to use
- Select a dataset preset for your species.
- Search for an RNA (e.g., sRNA
RyhBor CDSuvrA). - Tune filters: min io, min Of, and feature class.
- Click a partner to open globalMAP centered on that partner (rapid “drill‑around” exploration).
Tip: Sorting by Of surfaces the most specific targets first. Use the highlight box to track a short list of hypotheses while you refocus on different RNAs.
Tool guide: csMAP
What it shows: collapsed (comparative) interaction profiles so you can place multiple RNAs side‑by‑side and compare partner spectra.
How to use
- Select a species preset.
- Add two or more RNAs to compare (e.g., hub sRNAs or candidate sponges).
- Use differences in partner classes (5′UTR vs. CDS) to infer logic; copy candidate pairs to pairMAP for interface‑level inspection.
Tool guide: pairMAP
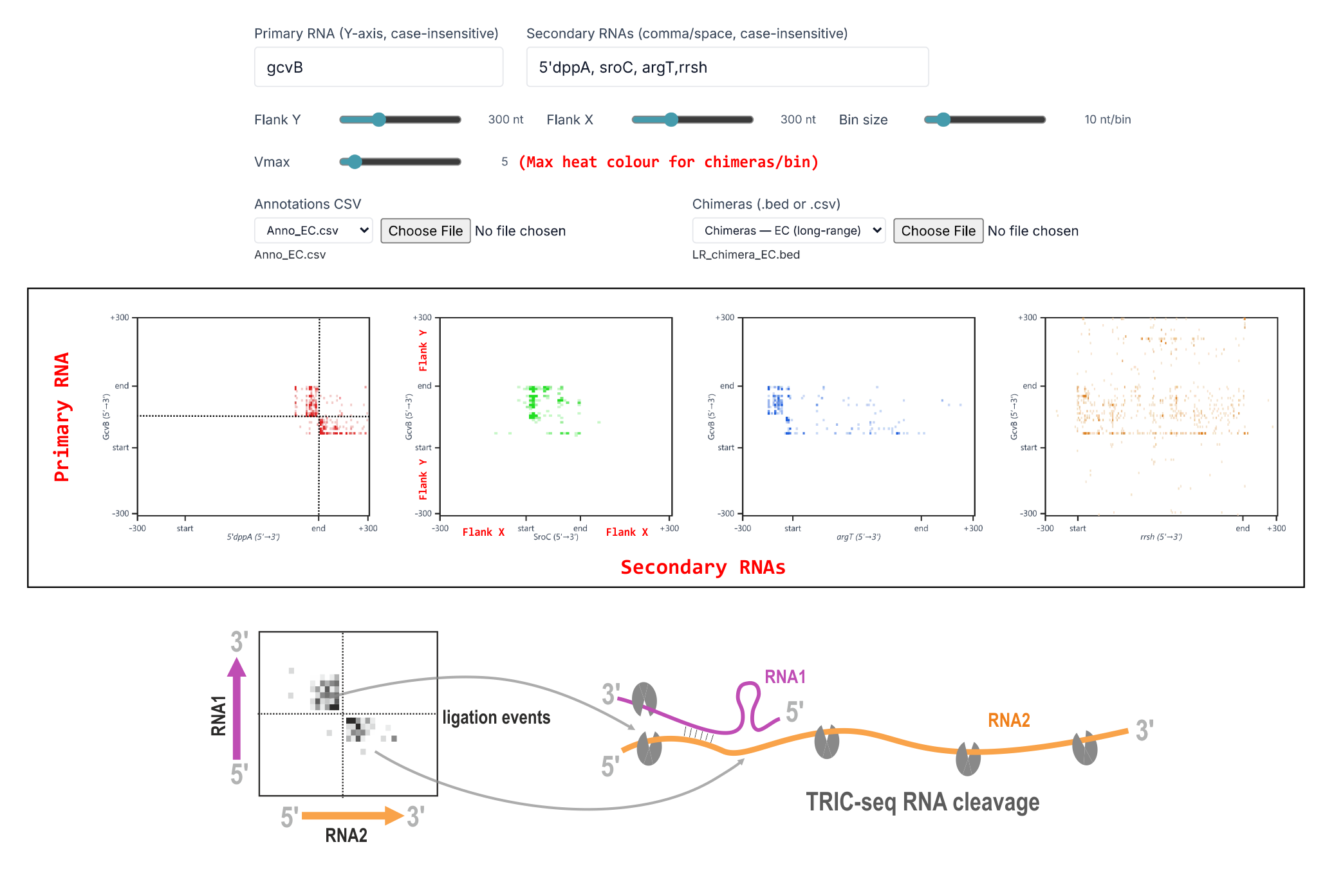
What it shows: a high‑resolution inter‑RNA heat map for a chosen pair (axes are nucleotide positions, binned).
How to use
- Enter a Primary RNA and one or more Secondary RNAs.
- Set Flank (±nt around each RNA) and Bin (nt/bin) for resolution.
- Interpret:
- Focused hotspot near diagonal → base‑pairing interface (typical sRNA–5′UTR).
- Diffuse CDS‑wide signal → co‑association/aggregation (e.g., mRNA–mRNA).
- 5′UTR hotspot + 3′UTR signal can reflect tertiary folding bringing ends together (not necessarily a second binding site).
Tool guide: foldMAP
What it shows: an intramolecular (self‑contact) map capturing an RNA’s average tertiary organization in vivo.
How to use
- Select a species and an RNA.
- Choose bin size to balance detail vs. sparsity.
- Look for domains (blocks along the diagonal), insulated boundaries, and interaction islands/deserts.
Note: foldMAP summarizes a population‑average contact map; multiple conformations can overlay into the observed pattern. Use the export buttons to save the contact map and long‑range profile (SVG/CSV).
